This license package contains the methods Hierarchical clustering, Hierarchical clustering with distance matrix, k-means clustering and Self-organizing maps
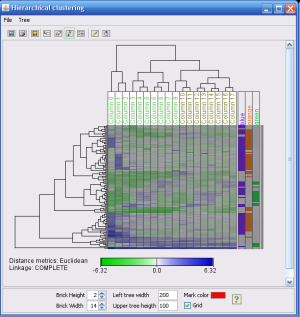
The Hierarchical clustering methods lets you choose between several distance metrics to order the genes based on similarity in their expression values. The image to the right shows a hierarchical clustering result window. Selections made in this window can be automatically forwarded to other J-Express windows to further explore for instance gene annotation or expression values.
The k-means clustering method divides the expression vectors into a predefined number of groups based on their expression values. This is a very quick way to separate the genes with the expression patterns you are looking for from the rest of the genes.
The Self-organizing maps method can produce results very similar to the k-means method, but in addition, the clusters are ordered in an intuitive way so that clusters with similar expression patterns are ordered together. There are also other analysis methods available for the SOM result, such as the value cell views that can be used for class prediction.
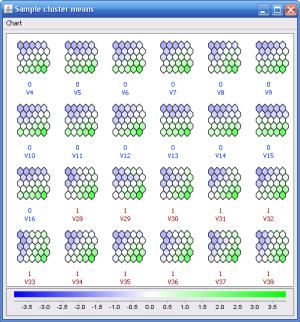
A SOM value cell view example
